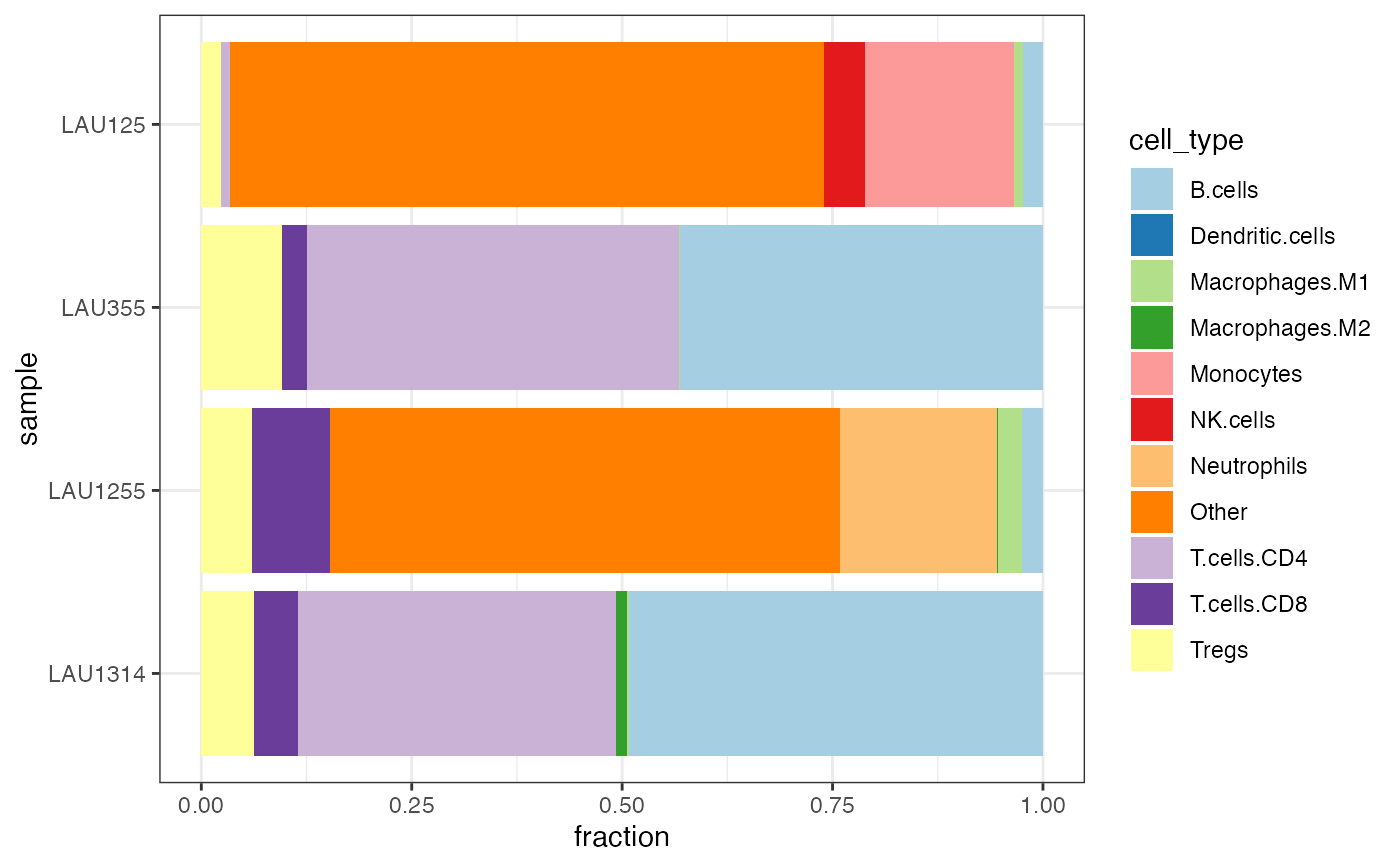
Plot the information on the tumor immune contexture
quantiplot.RdPlot the information on the tumor immune contexture, as extracted with
run_quantiseqr()
quantiplot(obj)Arguments
Value
A ggplot object
Examples
data(dataset_racle)
dim(dataset_racle$expr_mat)
#> [1] 32467 4
res_quantiseq_run <- quantiseqr::run_quantiseq(
expression_data = dataset_racle$expr_mat,
signature_matrix = "TIL10",
is_arraydata = FALSE,
is_tumordata = TRUE,
scale_mRNA = TRUE
)
#>
#> Running quanTIseq deconvolution module
#> Gene expression normalization and re-annotation (arrays: FALSE)
#> Removing 17 noisy genes
#> Removing 15 genes with high expression in tumors
#> Signature genes found in data set: 135/138 (97.83%)
#> Mixture deconvolution (method: lsei)
#> Deconvolution successful!
# using a SummarizedExperiment object
library("SummarizedExperiment")
se_racle <- SummarizedExperiment(
assays = List(
abundance = dataset_racle$expr_mat
),
colData = DataFrame(
SampleName = colnames(dataset_racle$expr_mat)
)
)
res_run_SE <- quantiseqr::run_quantiseq(
expression_data = se_racle,
signature_matrix = "TIL10",
is_arraydata = FALSE,
is_tumordata = TRUE,
scale_mRNA = TRUE
)
#>
#> Running quanTIseq deconvolution module
#> Gene expression normalization and re-annotation (arrays: FALSE)
#> Removing 17 noisy genes
#> Removing 15 genes with high expression in tumors
#> Signature genes found in data set: 135/138 (97.83%)
#> Mixture deconvolution (method: lsei)
#> Deconvolution successful!
quantiplot(res_quantiseq_run)
#> Warning: `aes_string()` was deprecated in ggplot2 3.0.0.
#> ℹ Please use tidy evaluation idioms with `aes()`.
#> ℹ See also `vignette("ggplot2-in-packages")` for more information.
#> ℹ The deprecated feature was likely used in the quantiseqr package.
#> Please report the issue to the authors.
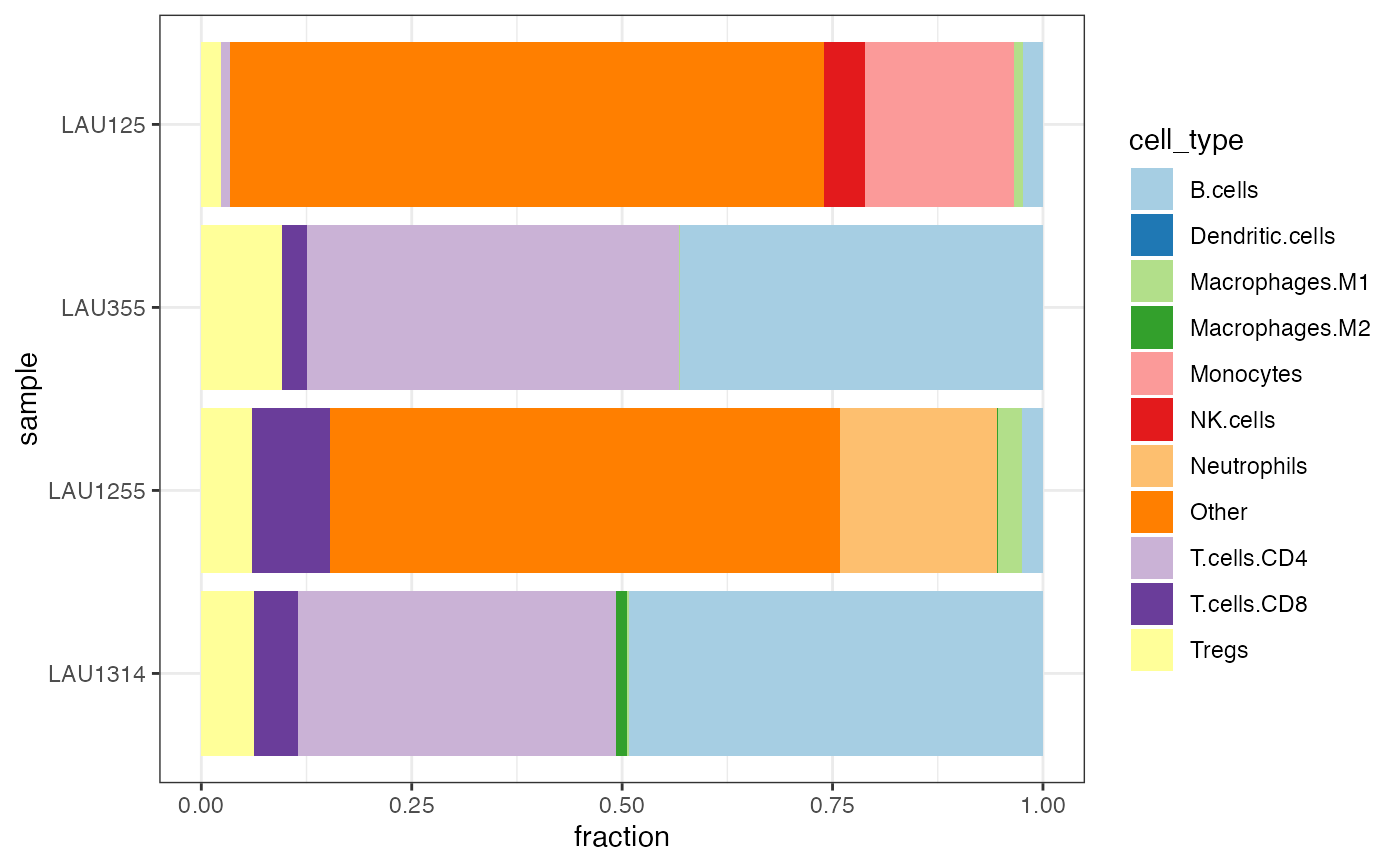 # equivalent to...
quantiplot(res_run_SE)
#> Found quantifications for the TIL10 signature...
# equivalent to...
quantiplot(res_run_SE)
#> Found quantifications for the TIL10 signature...