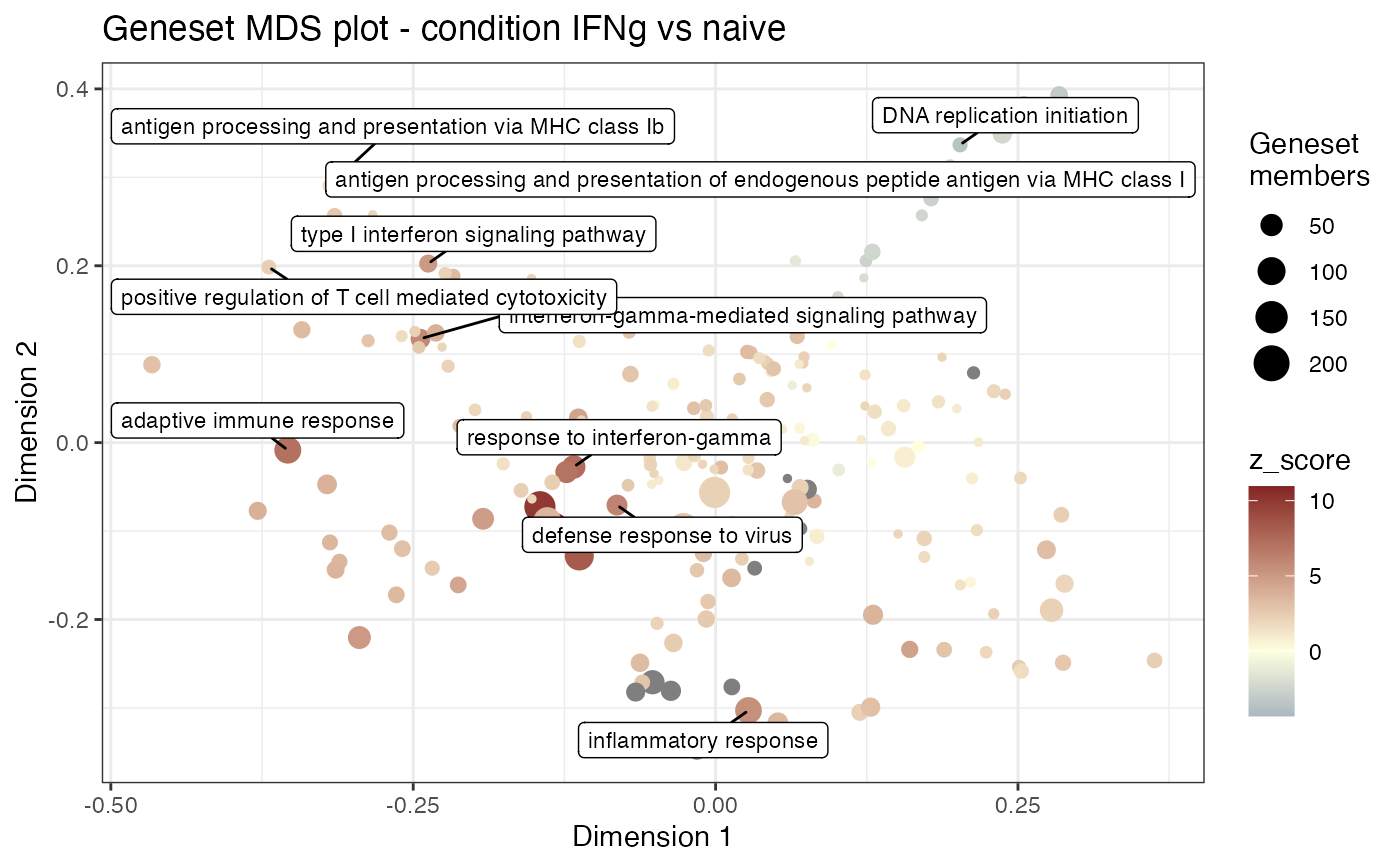
Multi Dimensional Scaling plot for gene sets, extracted from a res_enrich
object
gs_mds(
res_enrich,
res_de,
annotation_obj,
gtl = NULL,
n_gs = nrow(res_enrich),
gs_ids = NULL,
similarity_measure = "kappa_matrix",
mds_k = 2,
mds_labels = 0,
mds_colorby = "z_score",
gs_labels = NULL,
plot_title = NULL,
return_data = FALSE
)Arguments
- res_enrich
A
data.frameobject, storing the result of the functional enrichment analysis. See more in the main function,GeneTonic(), to check the formatting requirements (a minimal set of columns should be present).- res_de
A
DESeqResultsobject.- annotation_obj
A
data.frameobject with the feature annotation information, with at least two columns,gene_idandgene_name.- gtl
A
GeneTonic-list object, containing in its slots the arguments specified above:dds,res_de,res_enrich, andannotation_obj- the names of the list must be specified following the content they are expecting- n_gs
Integer value, corresponding to the maximal number of gene sets to be included (from the top ranked ones). Defaults to the number of rows of
res_enrich- gs_ids
Character vector, containing a subset of
gs_idas they are available inres_enrich. Lists the gene sets to be included, additionally to the ones specified vian_gs. Defaults to NULL.- similarity_measure
Character, currently defaults to
kappa_matrix, to specify how to compute the similarity measure between gene sets- mds_k
Integer value, number of dimensions to compute in the multi dimensional scaling procedure
- mds_labels
Integer, defines the number of labels to be plotted on top of the scatter plot for the provided gene sets.
- mds_colorby
Character specifying the column of
res_enrichto be used for coloring the plotted gene sets. Defaults sensibly toz_score.- gs_labels
Character vector, containing a subset of
gs_idas they are available inres_enrich. Lists the gene sets to be labeled.- plot_title
Character string, used as title for the plot. If left
NULL, it defaults to a general description of the plot and of the DE contrast- return_data
Logical, whether the function should just return the data.frame of the MDS coordinates, related to the original
res_enrichobject. Defaults to FALSE.
Value
A ggplot object
See also
create_kappa_matrix() is used to calculate the similarity between
gene sets
Examples
library("macrophage")
library("DESeq2")
library("org.Hs.eg.db")
library("AnnotationDbi")
# dds object
data("gse", package = "macrophage")
dds_macrophage <- DESeqDataSet(gse, design = ~ line + condition)
#> using counts and average transcript lengths from tximeta
rownames(dds_macrophage) <- substr(rownames(dds_macrophage), 1, 15)
dds_macrophage <- estimateSizeFactors(dds_macrophage)
#> using 'avgTxLength' from assays(dds), correcting for library size
# annotation object
anno_df <- data.frame(
gene_id = rownames(dds_macrophage),
gene_name = mapIds(org.Hs.eg.db,
keys = rownames(dds_macrophage),
column = "SYMBOL",
keytype = "ENSEMBL"
),
stringsAsFactors = FALSE,
row.names = rownames(dds_macrophage)
)
#> 'select()' returned 1:many mapping between keys and columns
# res object
data(res_de_macrophage, package = "GeneTonic")
res_de <- res_macrophage_IFNg_vs_naive
# res_enrich object
data(res_enrich_macrophage, package = "GeneTonic")
res_enrich <- shake_topGOtableResult(topgoDE_macrophage_IFNg_vs_naive)
#> Found 500 gene sets in `topGOtableResult` object.
#> Converting for usage in GeneTonic...
res_enrich <- get_aggrscores(res_enrich, res_de, anno_df)
gs_mds(res_enrich,
res_de,
anno_df,
n_gs = 200,
mds_labels = 10
)