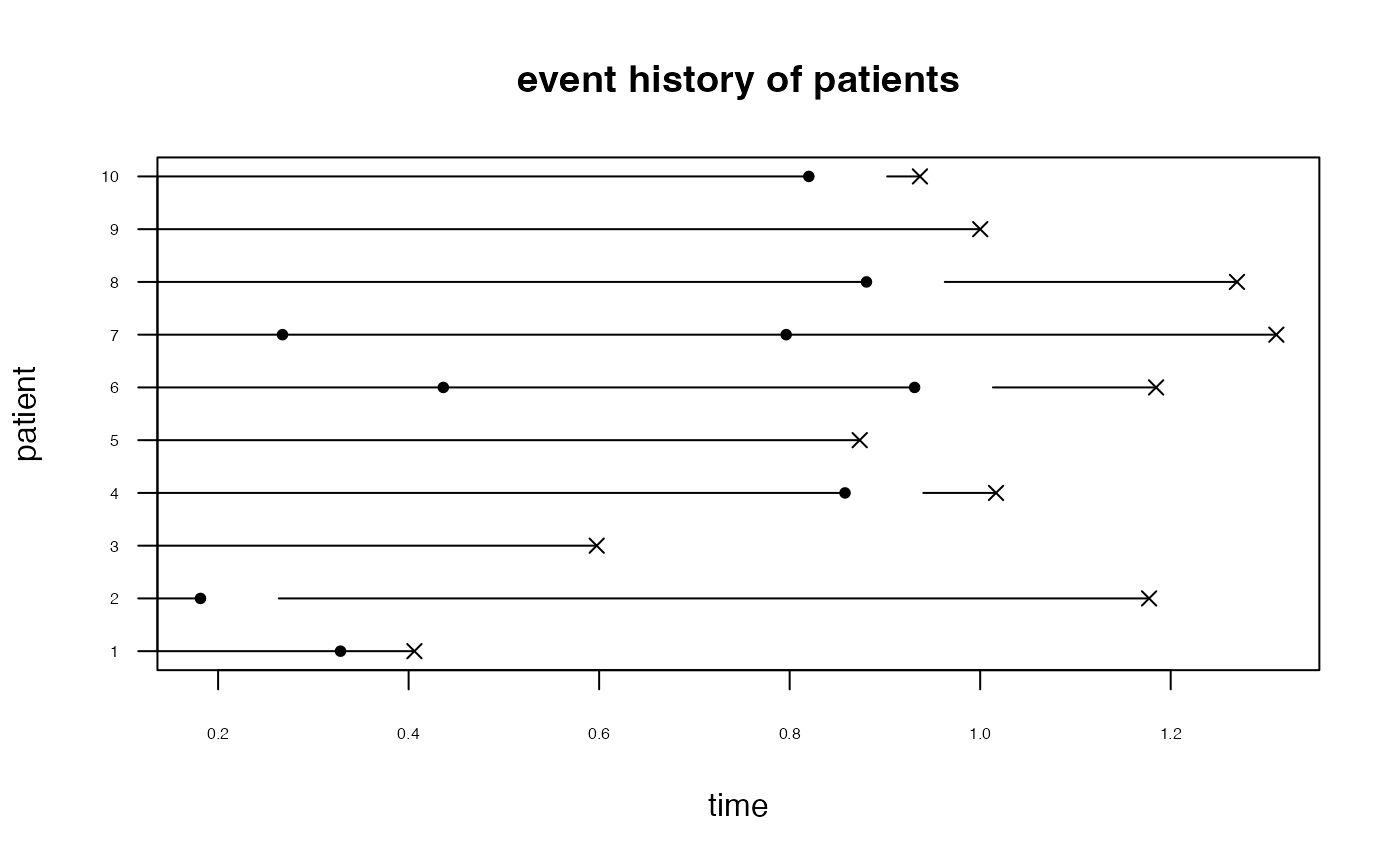
This function allows plotting of recurrent event data with a competing event.
simreccompPlot(
data,
id = "id",
start = "start",
stop = "stop",
status = "status"
)Arguments
- data
A data set of recurrent event data to be plotted. The input-data must include columns corresponding to:
id(patient-ID),start(= beginning of an interval where the patient is at risk for an event),stop(= end of the interval due to an event or censoring),status(= an indicator of the patient status atstopwith = 0 censoring, 1 = event, 2 = competing event)- id
the name of the
idcolumn, default is"id"- start
the name of the
startcolumn, default is"start"- stop
the name of the
stopcolumn, default is"stop"- status
the name of the
statuscolumn, default is"status"
Value
The output is a plot of the data with a bullet indicating a recurrent event, an x indicating the competing event and a circle indicating censoring.
See also
simrec, simreccomp, simrecPlot
Examples
### Example:
### First simulate a sample of 10 individuals (for more details see the help of `simreccomp`)
N <- 10
dist.x <- c("binomial", "normal")
par.x <- list(0.5, c(0, 1))
beta.xr <- c(0.3, 0.2)
beta.xc <- c(0.5, 0.25)
dist.zr <- "gamma"
par.zr <- 0.25
dist.zc <- "gamma"
par.zc <- 0.3
dist.rec <- "weibull"
par.rec <- c(1, 2)
dist.comp <- "weibull"
par.comp <- c(1, 2)
fu.min <- 2
fu.max <- 2
cens.prob <- 0.2
dfree <- 30 / 365
pfree <- 0.5
simdata <- simreccomp(
N = N, fu.min = fu.min, fu.max = fu.max, cens.prob = cens.prob,
dist.x = dist.x, par.x = par.x, beta.xr = beta.xr, beta.xc = beta.xc,
dist.zr = dist.zr, par.zr = par.zr, dist.zc = dist.zc, par.zc = par.zc,
dist.rec = dist.rec, par.rec = par.rec, dist.comp = dist.comp, par.comp = par.comp,
pfree = pfree, dfree = dfree
)
simreccompPlot(simdata)